hema.to Instructions for Use
User Manual release: 2025-01-21
Version 8
- (change to Version 1: chapters (1) and (2) added)
- change to Version 2: instruction to password reset and marker expression table added
- change to Version 3: updated diagnostic recommendation section
- change to Version 4: instructions for cell tree usage added
- change to Version 5: warning banners explanation added; SW lifetime added; further explanation on target population; disclaimer about "off-the-shelf" product
- change to Version 6 - including requirements from MDCG 2019-16 and updates based on hema.to_Intended Purpose and Classification_V08
- updated info on target population
- add analytical performance chapter
- added a separate warning section
- added a section on tec hnical description for using hema.to
- change to Version 7 - added 2-factor authentication
- change to Version 8- Manual cell annotation, updated label information and account settings interface
Note for the User
You will find all product information in the following three sections; (1) general information, which includes relevant IDs, numbers and identification marks, (2) device description, which includes intended purpose, use & user, functionality and contraindications, (3) instructions how to use app.hema.to and explanations of displayed items.
Can’t find what you’re looking for?
Contact support
Send a short e-mail (subject: help) to product@hema.to with a description of your question and your contact details. Alternatively, you can book a call with our Product Officer.
If it’s urgent, you can also call our support hotline directly (+49 177 2390536).
Report a bug or an incident
Send a short e-mail (subject: bug_report) to product@hema.to with a description of your problem and your contact details.
Send us your feedback
Send a short e-mail (subject: feedback) to product@hema.to with a description of your feedback and your contact details.
1. General information and product identification
1.1. Product Name
hema.to
1.2. Manufacturer details
Metzstr. 14B
81667 München
Germany
info@hema.to
VAT Number: DE348156268
EORI Number: DE461018766392877
National Trade Register: HRB270276
Actor SRN (EU): DE-MF-000029988
1.3 Shelf-Life / Expiry Date
Since hema.to pushes for software updates, every major/significant release is considered the end of life of the software and with it the old version of the software is decommissioned. This means that the shelf life of the cloud software is the time between two major software updates. All subsequent versions will be minor changes/releases.
2. Device Description
2.1. Intended purpose and function
hema.to is a stand alone software that supports physicians and laboratory staff in the analysis of immunophenotyping data stemming from cell suspensions of human specimens.
hema.to is a diagnostic aid for the screening, diagnosing and monitoring of hematological malignancies (B-NHLs).
2.2. Object of Measurement
hema.to supports the diagnostician in detecting hematological malignancies using immunophenotyping data stemming from flow cytometry measurements (so-called FCS or LMD files) of cell suspensions.
2.3. Intended Target Population
hema.to is a non-invasive diagnostic and screening aid using standard FC data generated by other medical devices; hema.to shall not impose any demographic restrictions like race or gender since diagnostic recommendations originating from flow cytometry (as stated by WHO) are independent of adulthood demographic characteristics in blood cancer malignancy detection. Pediatric cases shall be excluded from the analysis as childhood cancers are characterised by large variability.
2.4. Intended User
hema.to is intended to be used by personnel which have been trained to perform hematological analyses and diagnoses based on FC data. This could be either a physician and/or laboratory staff. Intended user does not have to have any extended knowledge about IT nor software engineering.
2.5. Intended Use
hema.to is used as a web application where standard flow cytometry (FC) data (FCS or LMD files) from cell suspension measurements (e.g. from blood or bone marrow) can be uploaded, to efficiently support its user in making quantified differential diagnosis and accompanying clinical reasoning.
Since hema.to is software based on industry-standard FC data, it is non-invasive and can be used anywhere, anytime, for any desired duration, as often as is desired. No hardware or software upgrades, hardware security measures, additional accessories or other adaptations of the FC hardware are required. hema.to does not require exclusivity in FC data analysis; it does not alter nor delete original data.
Medical Conditions and Medical Indications
hema.to is intended to be used with patients who are suspected of a hematological malignancy, and for whom flow is part of standard diagnostics. Indications are FC data from cell suspensions (including but not limited to blood or bone marrow).
Contraindications
There are no contraindications in addition to those already imposed by the (normal) use of FC.
Compatibilities
hema.to can only be used in combination with flow cytometers that produce standard FCS or LMD files, which are processed on their computer in their internet browser of choice.
2.6. Test principle
hema.to uses supervised machine learning (ML). The “supervision” in this case are the malignancy sub-type labels, and the algorithm learns the relationship between the flow data (input) and the malignancy sub-type (output). That is one reason why we ask for your diagnostic feedback — by doing so, hema.to’s diagnostic recommendations improve over time. This will make future diagnoses quicker and better, improving both your workflow and clinical decision making.
Our ML algorithms use deep learning.
For technical details regarding Principles of Operation please contact info@hema.to.
2.7. Performance characteristics
2.7.1 Analytical Performance characteristic
Analytical performance indicators of hema.to include:
- Confidentiality:
- confidential handling of patient data is ensured
- Integrity:
- 1. FC data, upon transfer to hema.to, are notcorrupted nor destroyed by the software. Files are not manipulated at their original location on customers servers.
- Reliability:
- all functionalities work reliably
- Generalisability:
- This medical software is not an of-the-shelf product, it requires pre-training for each new customer or each new significant change in the laboratory standard operating procedures (among others: antibody panel, compensation settings, flow cytometer characteristics which can be seen in the data)
- If unknown data is uploaded into hema.to safety measure will by applied (see ‘4. Warnings’)
- Usability engineering:
- performed according to usability engineering standard EN 62366-1.
- Data quality:
- File format: FC data: .lmd or .fcs
- Data integrity: Stored data has the expected number of entries. Computed file hashsum agrees with value stored in file, if included.
- Consistency of the measured antibody panel: Antibody panel in file agrees with one of the allowed panels.
- Number of events: ≥ 6000 recorded events.
2.7.2 Clinical Performance characteristic
The clinical performance indicators of the hema.to include:
- Diagnostic Accuracy
- diagnostic sensitivity of ≥ 90%
- diagnostic specificity of ≥ 85%
- positive predictive value of ≥ 75%
- negative predictive value of ≥ 90%
- positive likelihood ratio of ≥ 5
- negative likelihood ratio of ≤ 0.1
- Diagnosis Time ≤ half the time of manual diagnosis
Disclaimers
- We do not replace medical judgment. hema.to is not intended to replace the diagnosticians’ best judgment, nor to deliver fully automated diagnoses.
- Time is not reduced to zero. hema.to generates an average time reduction (cf. the clinical claims), but time reduction may vary, depending on whether the user chooses to increase e.g. discussion time for some cases as compared to their normal workflow. Moreover, hema.to cannot fully automate the diagnostic process — the expert user will still have to operate hema.to for a minimum amount of time per case.
- This product is not so-called ‘off-the-shelf’ product. It requires fine-tuning the model in order to provide diagnostic recommendation for every new customer or after each significant change in laboratory standard operating procedures. Analytical requirements can be found in Performance Evaluation Repot (PER_2023_V02).
Risks & Side-effects
Since hema.to does not perform medical interventions (it is non-invasive diagnostic software), hema.to does not cause side effects, either positive or adverse. hema.to does not require exclusivity; flow cytometry data can be analysed using any other software; hema.to does not manipulate on raw data at any point.
3. Using hema.to
3.1. Personal account
You should have received your hema.to log-in credentials from us. You can log-out and log back in to hema.to with these credentials. Keep these credentials secure, and do not pass them on to others. URL for log-in: app.hema.to
Upon entering your username and password, you will be prompted to complete a two-factor authentication (2FA) process for added security. A verification code will be sent to the email address associated with your hema.to account. Please note that this verification code is only valid for 5 minutes from the time the email was received. If the verification code expires, you can initiate the login process again to receive a new one.
You can reset your password via the forgot your password button. Please provide the email address that is associated with your hema.to account and proceed with guidance received in the automatic email. Please contact product@hema.to if you did not the receive email notification.
If you would like to make a new account for a colleague, send us an email (product@hema.to, subject: account).
If you have lost your credentials, contact hema.to support via e-mail (product@hema.to, subject: account) or book a call. If it’s urgent, you can also call our support hotline directly (+49 177 2390536).
Managing accounts
If you are the “admin” of your organisation, you can manage the accounts within the account settings. You will be able to create, deactivate and edit existing accounts.
To add a new account, you would need to provide the new user’s details (name, email…) and once the account is created, the new user will receive a verification email to start the login workflow.
3.2. Uploading flow data
Uploading flow data is easy. Both FCS and LMD files are accepted.
At the moment, we only work with laboratories where we have demonstrated a reliability comparable to (or exceeding that) of industry-standard diagnoses. That means that you won’t be able to upload files with arbitrary panels at this point.
If there was something wrong with a file, you’ll see a warning or error message. Things that can be wrong include: not a FCS or LMD file, corrupted file, a file that does not conform to the FCS standard. Moreover, we reject files where we cannot anonymize the file name. This can happen if the name of the file does not correspond with the naming scheme sent to us in sample data.
In these cases, hema.to tries to show a clear warning or error message, explaining the problem.
Please note, a change in the laboratory's internal standard operating procedures* that leads to a change in the (FCS or LMD) data structure or the statistical properties must be reported to hema.to as soon as possible. Modified datasets must not be uploaded via the platform without prior agreement with hema.to.
* In particular, but not limited to, a change in:
- the naming convention of the FCS/LMD files,
- the storage location and type of personal information in the FCS/LMD,
- the naming scheme of the antibodies and fluorochromes of the panels in the FCS/LMD files, and also
- a change in the flow cytometers used,
- panels,
- as well as the staining process.
Uploading flow data in bulk mode
You can also upload multiple cases at once. The files are automatically matched by sequence ID based on the filenames.
You can start analyzing cases before all files have finished uploading.
You can see that a case is ready to be diagnosed by the label “Click to perform diagnosis”:
Please note, uploading many files may take a few minutes.
3.3. Clinical indications
hema.to’s clinical indications of a case in “Case Data” consist of cytograms and case abnormalities visualized in those cytograms.
The cytograms display all cells in the flow data, with the B-lymphocytes colored differently from other cells by means of gating (also indicated in the Case Data panel).
The abnormalities consist of overpopulated and underpopulated cell populations (colored in blue and red, respectively), as compared to an average healthy phenotype. These colorings give a rapid and reliable indication of the expression pattern.
Be sure to always briefly check the validity of the B-cell gate by CD19 positivity, especially if the results appear aberrant. If the B-cell gate does not appear correct, we advise to resort to manual diagnosis.
Gating
No manual gating is necessary in order to use hema.to. Our single-cell classification algorithms and plotting service identify and represent the corresponding cell populations in each tube.
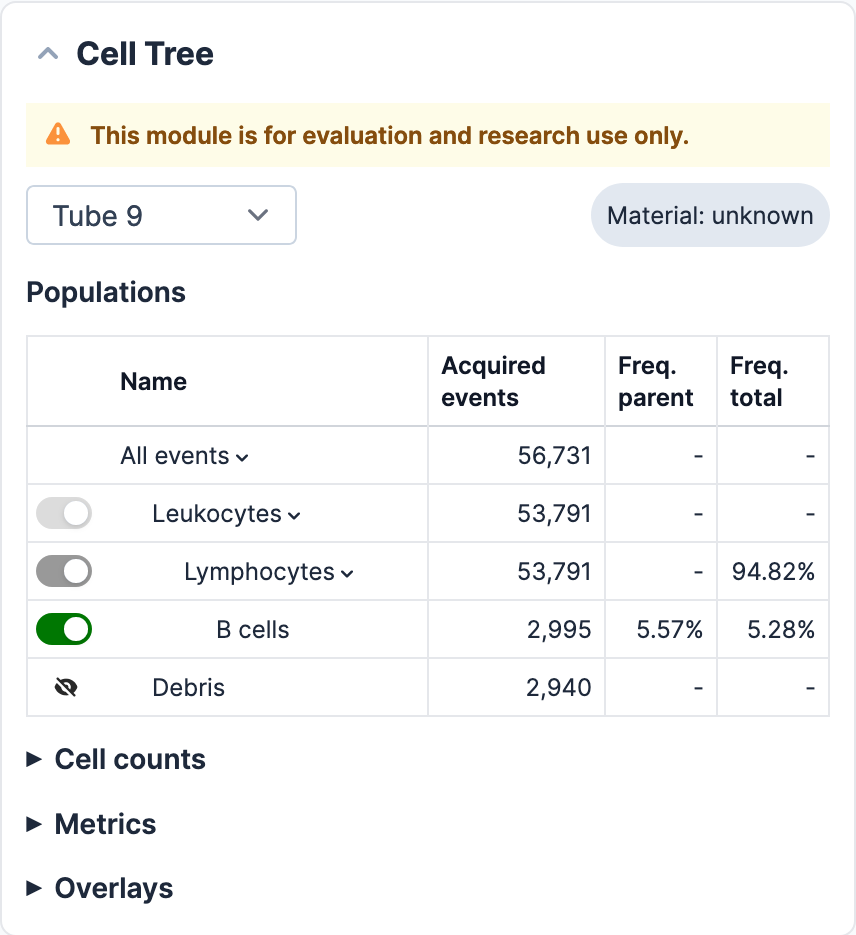
Cell Tree & interactive population visualisation
Cell populations that are displayed in the cytograms can be visualised in the adjacent Cell Tree given their assigned colour. Clinically-relevant cell populations are highlighted in bright colours, whereas other cell types and artefacts, are displayed in non-intrusive colours. Each cytogram can be enlarged to allow the user to take a closer look at the displayed cell populations.
In the Cell Tree, the hierarchical relationships between cell types are shown. In addition, for each population that appears in the cytograms, corresponding mathematical calculations are also displayed.
The visualisation of each cell population displayed in the Cell Tree can be enabled or disabled by the user by clicking on the button next to each population. When a population is enabled, it will appear highlighted in the corresponding cytograms in its given colour. For a population to be enabled, all parent populations have to be enabled as well. In the case a parent population is disabled and then enabled again, daughter populations will preserve their previous status (example: B cells are disabled but TNK and other lymphocytes are enabled ➡️ lymphocytes are disabled ➡️ lymphocytes are enabled again ➡️ B cells remain disabled and TNK and other lymphocytes enabled). There is, however, an option, to reset all changes and go back to the default configuration by clicking on “reset”.
Manual cell annotation corrections
In case you do not agree with the cell classification predictions shown in the cytograms or would like to adjust the selections, there is a manual option to do so. To modify a pre-existing selection, the corresponding cytograms needs to be enlarged. Once enlarged, the “lasso” tool icon needs to be selected to active the possibility to do manual annotations. Once the lasso tool is active, user can draw a selection area to cover the cells of interest. After cell selection has been made, it is possible to assign the selection to one of the available cell populations in the Cell Tree by interacting with the dropdown menu. By assigning the select cells to a population and clicking “apply”, the corresponding cells will appear highlighted in the corresponding colour in all other cytograms where they are displayed.
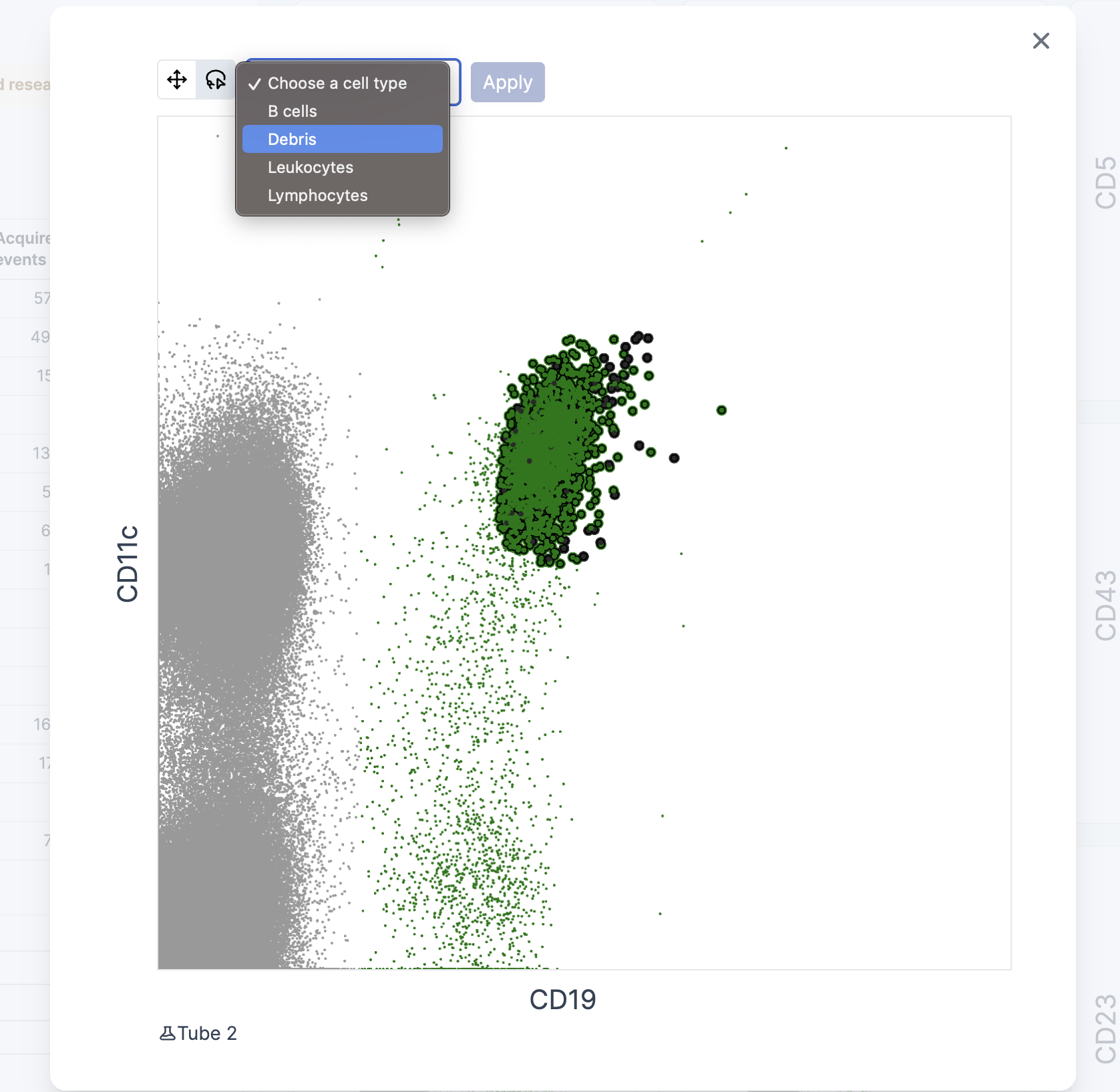
It is possible to make as many changes as desired, including changes made in different cytograms.
After all changes have been made, it is necessary to save or discard these changes. As a reminder, the following pop up window will appear at the bottom of the page.
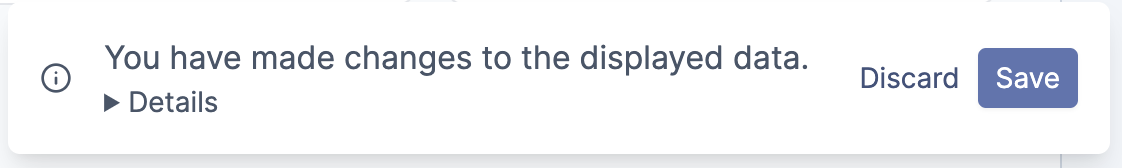
Changes made to populations will be reflected in the Cell Tree and doctor letter numerical calculations.
Manual adjustment of quadrants
Corrections to the thresholds in the cytograms (kappa/lambda plot) are possible through manual adjustment of the quadrant in the cytograms. You should enlarge the cytogram and move the lines by dragging them with the cursor. Make sure you update the gating by selecting the corresponding button; select Reset, if you want to discard your changes.
This is of interest, for example, to update the kappa/lambda ratio if you do not agree with the predictions made by our software.
Changes made through these changes will be reflected both in the Tube metrics and in the marker expression table on the “diagnosis” tab.
Tube metrics
Metrics for each tube are displayed in the corresponding section (next to the cytograms). Numbers which can be found there include: Kappa (K+) and Lambda (L+) - cells (relative and absolute); Kappa/Lambda (K/L) ratio, as well as CD4/CD8 T cell ratio.
Cell counts manual input
For the calculation of absolute cell counts, values for “white blood cells (cells/μL)” (wbc) and “lymphocytes” (cells/μL) can be manually entered. When values are saved, two additional columns are displayed on the cell tree, count (cells/μL) and Freq. wbc, which include the corresponding calculations for each cell population.
The “lymphocytes” input box was implemented for cases where not all leukocytes are acquired in the FCS file (as it is depicted in the FS-A/SS-A plot below). This alternative helps prevent miscalculations. In such cases, it is preferable that the “lymphocytes” count is entered and used for the calculation.
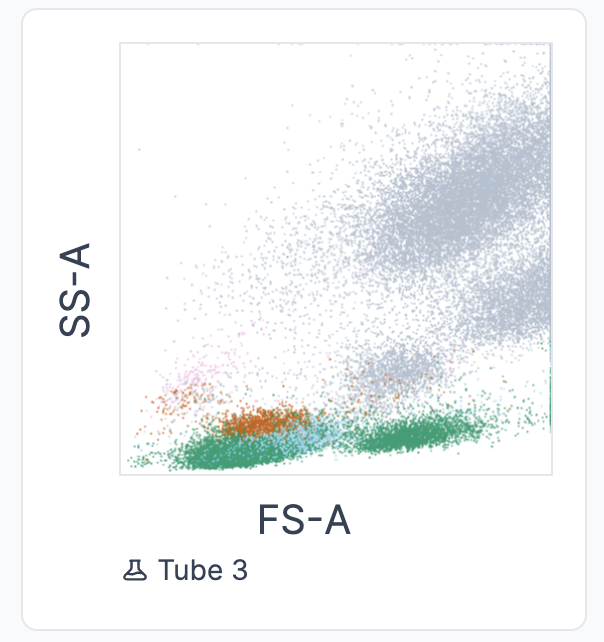
In cases where both numbers (”white blood cells” and “lymphocytes”) are available, only the “lymphocytes” will be used for the calculations to prevent the above-mentioned issue. In this cases, the following message appears:
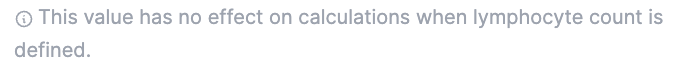
3.4. Diagnostic recommendations
Our diagnostic recommendations follow expert consensus. The confidences indicated represent how sure hema.to is of a gold standard diagnosis. For example, if the indicated confidence for a case is 95% for Hairy Cell Leukemia, then about 95% of experts would diagnose this patient with Hairy Cell Leukemia. We give such confidence scores for the malignancies:
- HCL (Hairy Cell Leukemia)
- CLL/MBL (Chronic Lymphocytic Leukemia/Monoclonal B-Cell Lymphocytosis)
- PL (Prolymphocytic Leukemia)
- FL (Follicular Lymphoma)
- MCL (Mantle Cell Lymphoma)
- LPL (Lymphoplasmacytic Lymphoma)
- MZL (Marginal Zone Lymphoma)
- normal (non-pathological)
If a diagnostic recommendation is available for a case, it will be shown as follows on the case page:

3.4.1 Marker expression table
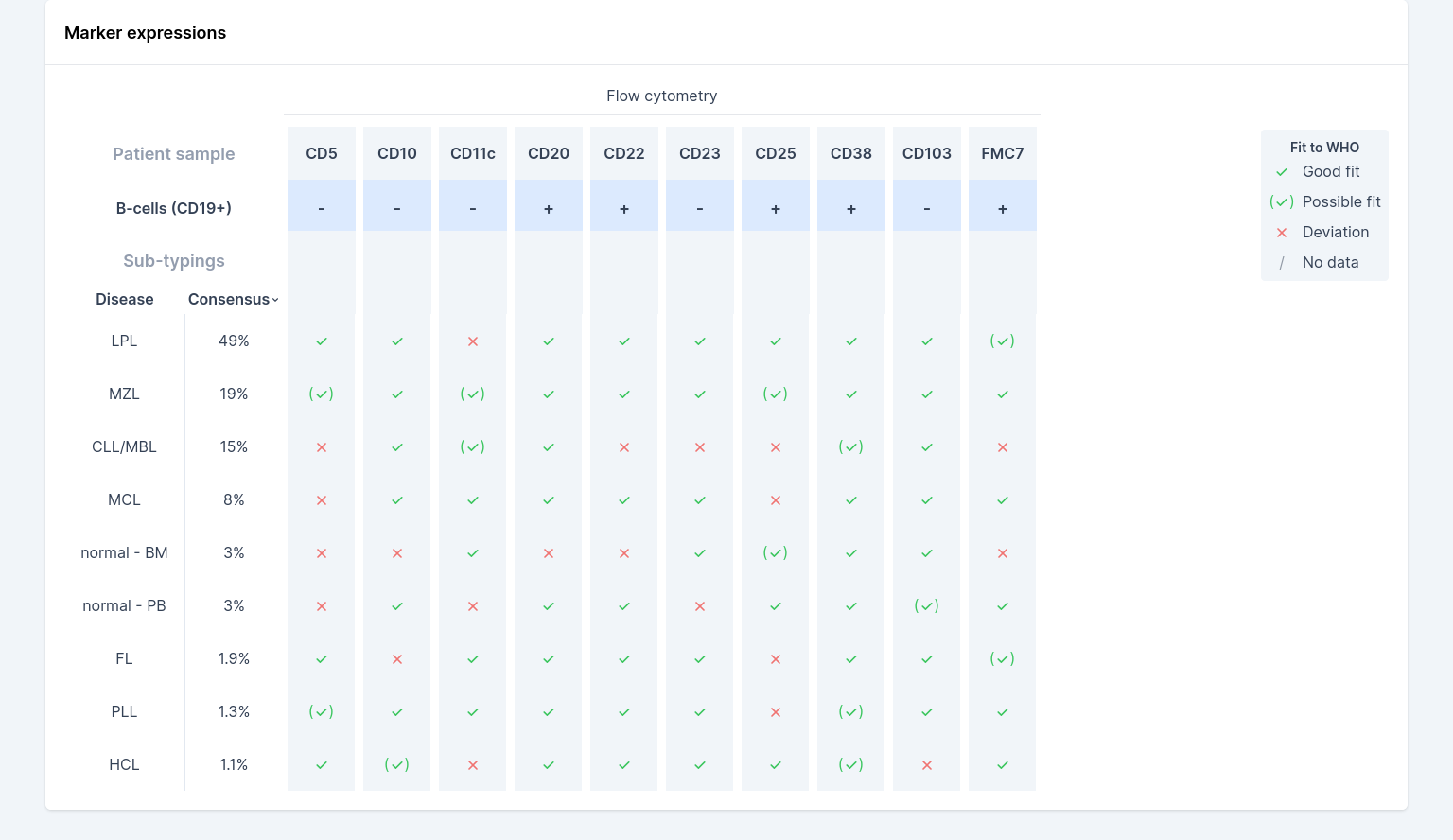
Expressed markers are measured directly from the FCS/LMD file and then compared to the standard WHO diagnostic criteria. Measured markers (blue row, see screenshot) are either negative (”-”) or positive (”+”). Marker expression is measured by locating the two largest clusters of cells (from all lymphocytes), and then checking whether the number of events in the positive cluster exceeds a lab-configured threshold. You can see the ratio of B-cells with positive expression, relative to all B-cells, by hovering over each marker column.
The markers from the sample are then compared to the WHO diagnostic criteria per sub-type (table section: ”Sub-typings”); a green check-mark indicates a good fit, green-check mark in parentheses indicates a possible fit and a red cross indicates a mismatch to the WHO expectation for that sub-type and marker combination (a gray slash indicates that there is no WHO expectation available for this marker and sub-type combination). This table can be used to assess to which WHO sub-type the patient sample matches best. However, these criteria will not unambiguously point to the correct sub-type and are only clinical insights and should always be combined with your expert medical judgment.
Finally, the “Consensus” column indicates hema.to’s confidence in expert consensus for that sub-type (see 3.4. Diagnostic recommendations). Because often the WHO isn’t unambiguous as to sub-type, hema.to’s consensus estimate takes into account the full FCS data using machine learning models trained on thousands of expert-annotated samples.
If multiple classes are all seen as approximately likely, this could be an indication that the gating was sub-optimal (see also the chapter on “Clinical Indications” in this manual), the sample is of low quality, that the patient is undergoing therapy, the patient suffers from a malignancy not included in the list, or other abnormalities are present in the data.
The above screenshot is an example of diagnostic recommendations that are not highly certain: for this example, extra attention should be given to the diagnosis, since hema.to wasn’t able to give high-quality recommendations.
3.4.2. Diagnostic recommendations - Acute Leukemia module
In addition to diagnostic recommendations for BNHLs, hema.to included an Acute Leukemia module into the app. As of now, this module is for research use only. To make this clear, you should be able to see the warning tab:

Once you open a case, if predictions are available for both modules, you can change views from one model to the other one by clicking on the toggle at the top of the page. In the Acute Leukemia model, we support diagnostic recommendations for Acute Myeloid Leukemia (AML) and Acute Lymphoblastic Leukemia (B-ALL). However, since this module is for research purposes only, you should confirm that you acknowledge the nature and limitations of the module before approving a diagnosis.
3.5. Approving a case
You first have to select your own diagnosis, before you can approve the case. You can select more than one malignancy, if you are not sure. Moreover, you can select additional methods based on which your diagnosis has been made (in addition to Flow cytometry results).
A case will be designated as “unapproved” if you haven’t approved it yet. You can approve a case at any later point, or change the malignancy you assigned to it.
You may also add case notes for recording your own diagnostic reasoning. These notes are stored in a non-editable log, so that you can always come back later to see what your or your colleagues’ diagnostic reasoning was, as in the screenshot below:
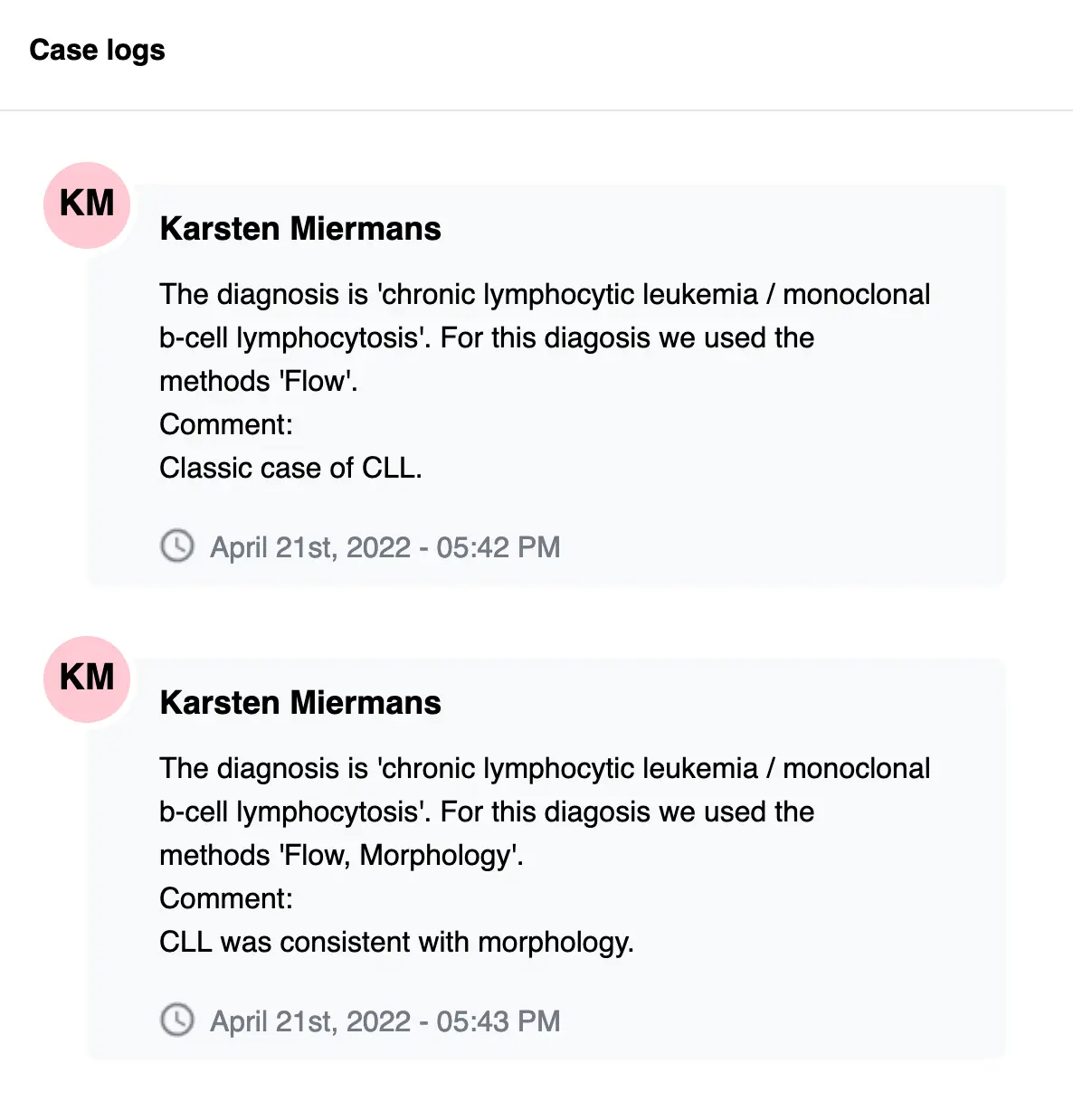
A case will be designated as “unapproved” if you haven’t approved it yet. You can approve a case at any later point, or change the malignancy you assigned to it.
Please note, in order to approve diagnosis you require such permission rights for your hema.to account.
Case recall
At any point, you can go back to your list of (un)approved cases. You can re-examine previously approved cases, or approve cases you haven’t approved yet.
Unapproved cases are under the “Your Active Cases” heading and contain a note that reads “Click to perform diagnosis”. Approved cases are under the “Approved Cases” heading. You can make changes to approved cases, see the “Approving a case” section of this manual.
4. Warnings
4.1. Offline banner

4.2. Quality Control

4.2.1. B-cell depletion algorithm

4.2.2. Low-confidence warning

4.3. Warning banner - lower accuracy upon integration

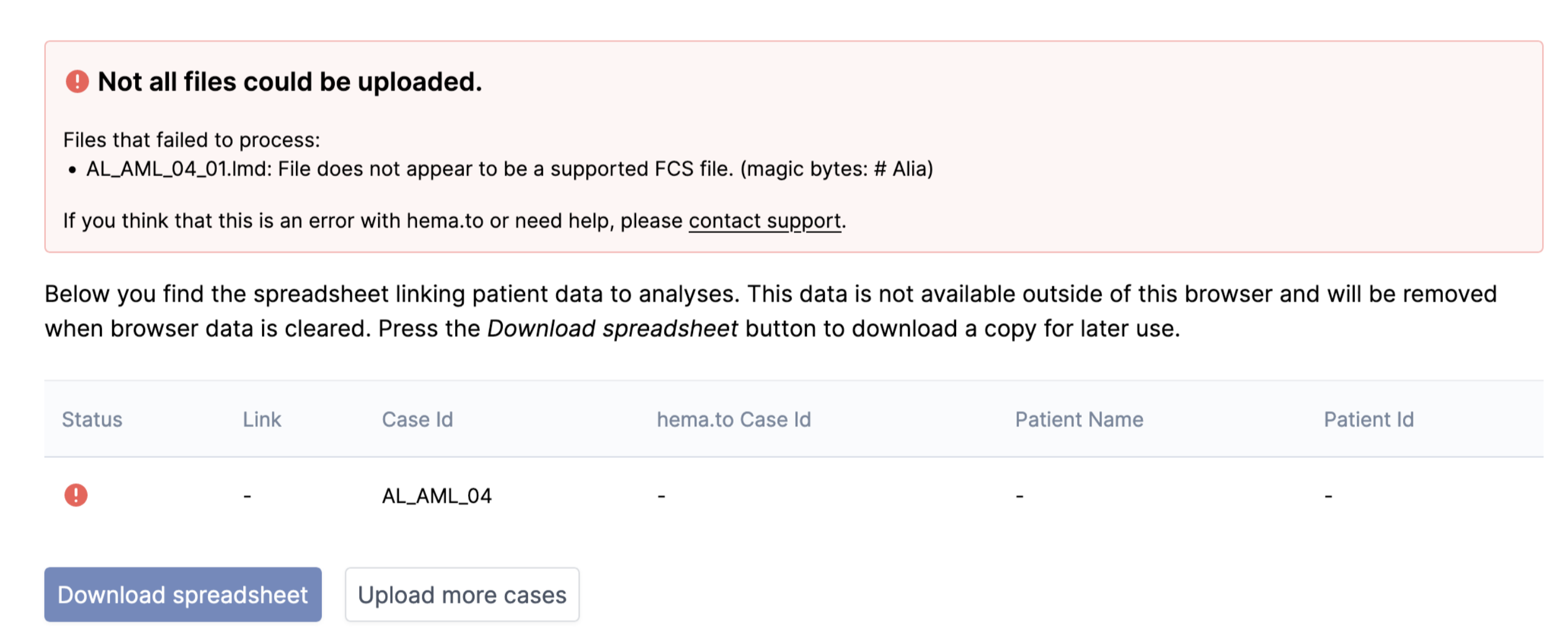
4.4. Flow parsing errors show in front-end
4.5. IVD under investigation

4.6. Abnormalities could not be visualised for all marker combinations

4.7. Results are not available
4.7.1. Diagnostic recommendations are not available

4.7.2. Cytograms could not be generated


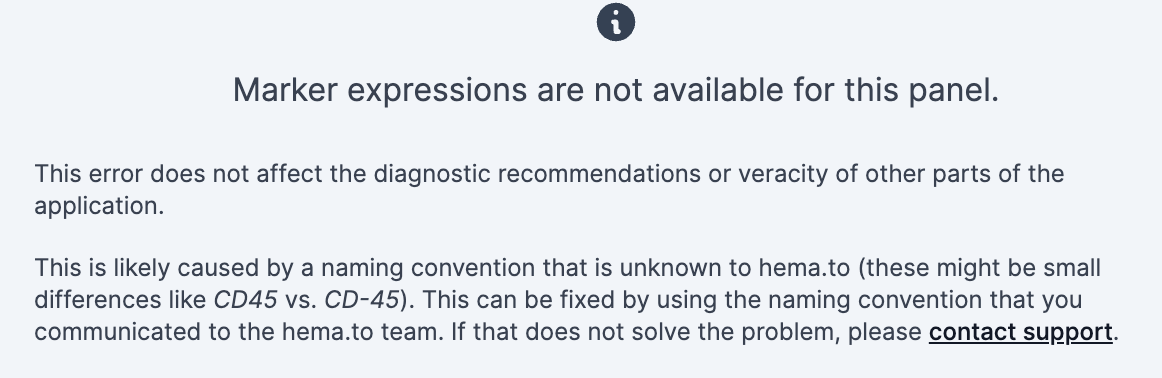
4.7.3. Marker expressions are not available

4.15. Analysis in progress

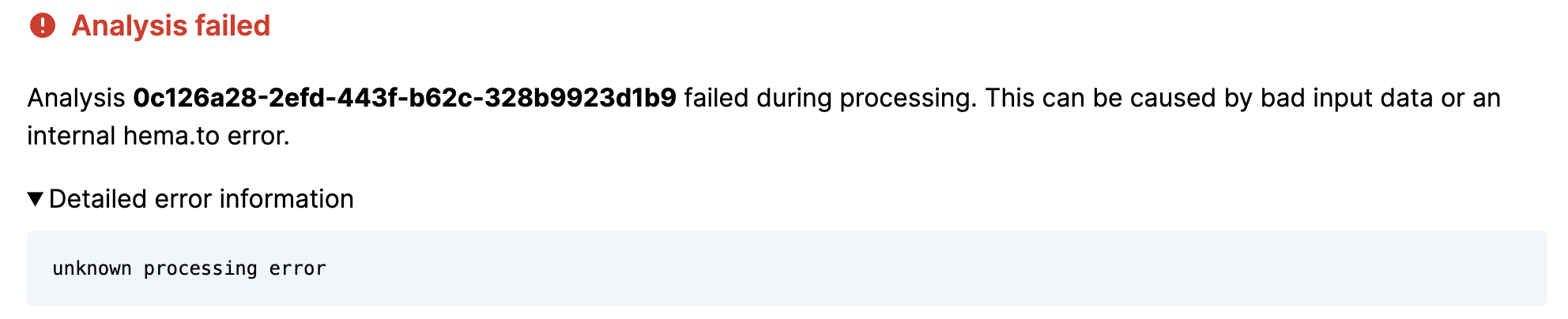
4.16. Analysis failed


4.17. Absolute cell counts not displayed

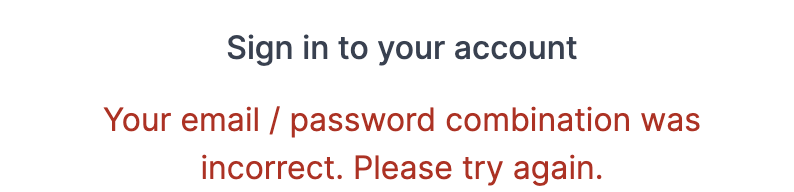
4.18. Sign in error
4.19. Cell tree research only use
5. Technical Description (incl. IT security) for using hema.to
5.1 Technical Description
The hema.to product, a cloud-based web application for diagnosing blood cancer, integrates comprehensive security measures to ensure its safe and secure operation, transport, storage, and use.
Response to Security Failures:
In case of a detected failure to maintain security, we immediately alert system administrators and initiating containment measures to prevent further unauthorized access or data breaches. The impact of such a failure on patient care, data integrity, and clinical workflow is minimized through these rapid response mechanisms. The system is designed to ensure that patient data remains confidential and secure, and that the integrity of the diagnostic process is not compromised.
Technical Security Measures:
- Configuration and Infrastructure:
- Network Security and Cloud Infrastructure: Our cloud servers, running the application, are configured to use only essential network ports and services, reducing potential vulnerabilities.
- Software:
- Data Encryption: All data transmitted to and from the user's browser is encrypted using SSL/TLS protocols. Additionally, sensitive data stored on our servers is encrypted at rest.
- Browser Security: The application leverages browser-based security features, including cookie attributes and CSP, to prevent XSS attacks and other web security risks.
- Password Management: Users are required to set a new, strong password upon first login, and have the option to change their password through the settings page.
- Operations:
- User Authentication and Session Management: The application employs strong authentication and session management controls. Sessions are time-limited and terminated securely after logout or periods of inactivity.
- Logging and Auditing: The software maintains logs of user interactions for each case, detailing actions and the responsible user. These logs are accessible through the case details page for transparency and accountability.
- Patch Management: Regular updates are applied to the software and its components to address vulnerabilities and maintain security.
Impact on Patient Care and Clinical Workflow:
The security features of our software are designed to operate seamlessly, ensuring that the diagnostic process is not hindered by security measures. The encryption of data, both in transit and at rest, ensures the confidentiality and integrity of patient data. The logging and auditing capabilities provide a detailed trail of user actions, vital for clinical accountability and traceability.
5.2. Basic Information
| IT and Hardware Requirements / Characteristic | Information |
|---|---|
| Computer Access | A computer will be needed to access the product. We strongly advise to not leave the computer unattended; especially when using hema.to. |
| Internet Access | We strongly advise to use a secure internet connection |
| Browser | All evergreen browsers of choice (two last versions) |
| Account Management | See point 3.1. Personal account |
| Session Management | We strongly advise you to log out of hema.to manually when used on shared computers. Otherwise, the session will be terminated after 30 days of inactivity. |
| Antivirus/Malware | follow best practices |
| Firewall | follow best practices |
5.3 Risk profile of the medical device and the corresponding IT security objectives
Protection of sensitive information: this device process raw flow cytometry files that cannot be used as the source to identify individual patients. We do not collect any sensitive information (e.g patient ID, patient name, patient address), furthermore, all our analysed data is anonymised.
Our servers are located in the European Union and protected from external access.
Requirement for uninterrupted operation: It is required to have internet access for the device to fulfil its intended purpose.
